Overview
My lab researches sex-specific selection and other context-specific selection, where contexts could be alternative tissues, life stages, environments, etc. Such selection can generate genetic tradeoffs known by familiar terms such as sexually antagonistic selection, antagonistic pleiotropy, ontogenetic conflict, or gene-by-environmental interactions. These phenomena have important evolutionary implications regarding the maintenance of genetic variation in fitness, local adaptation, and population viability, but can also have important applied value to contemporary issues in human society, such as wildlife conservation, pest management, and genetic disease. Past research has focused on sexually antagonistic genetic variation using quantitative genetics, experimental evolution, and transcriptomics. Future work will expand this work toward other forms of genetic tradeoffs (mentioned above) using a broader set of methodologies including genomics, ATAC-seq, CRISPR, and mathematical modeling.
Informal inquiries about joining the lab are always welcome. Get in touch!
Detecting antagonistic loci
Despite a rich quantitative genetic understanding of sexually antagonistic genetic variation, we know little about how many such polymorphisms are in the genome, whether they tend to be protein coding or regulatory, what traits they underly, and what their molecular and population genetic properties are. Past work used experimental evolution to force alternative male- and female-benefit alleles onto oppositely marked chromosome pools followed by genomic and/or transcriptomic contrasts of said pools to identify regions of the genome under sexually antagonistic selection. That remains a powerful tool. Future work will aim to develop novel methodologies for identifying candidate antagonistic polymorphisms (including those oppositely selected in opposite sexes, tissues, life stages, environments, etc.) using a combination of allele-specific expression, ATAC-seq, CRISPR genome editing, experimental evolution, and mathematical modeling. Much of this work aims to leverage a conspicuous pattern of Mendelian inheritance that antagonistic polymorphisms should be enriched for: dominance reversal (see next section).
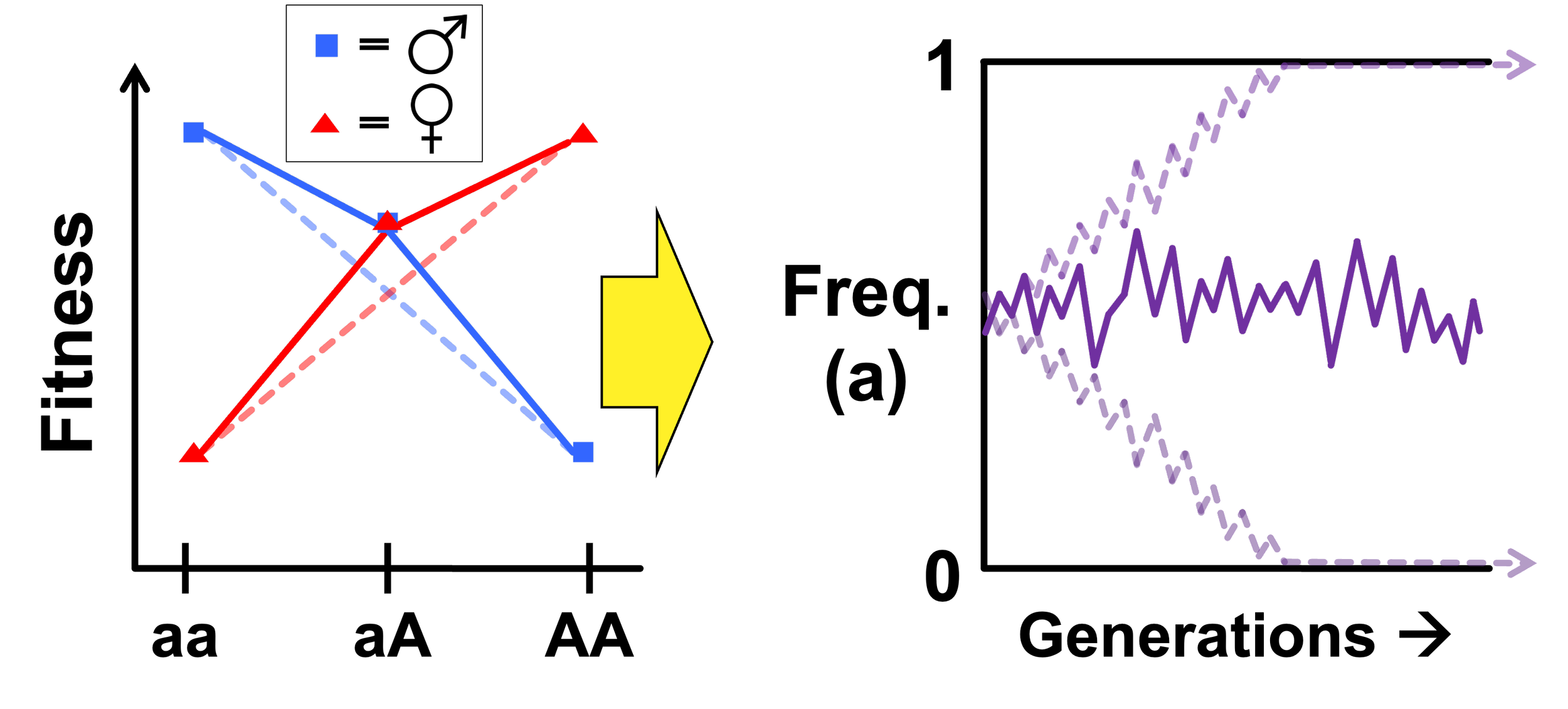
Sex-specific dominance reversal
Sex-specific dominance reversal - where the allele that benefits a given sex is also dominant in that sex - can partially resolve genetic conflicts, facilitating the maintenance of genetic variation harboring stored adaptive potential. Dominance reversals under antagonistic selection and have seen growing theoretical and empirical support. Thus, understanding how exactly these sex-specific dominance reversals occur may prove crucial to our ability to detect antagonistic polymorphisms.
Sex differences in (allele-specific) gene expression brought on by certain combinations of sex-limited trans-acting proteins, their concentrations, and/or cis-acting SNPs in their regulatory regions may be a common means by which sex-specific dominance reversal is achieved. We have built forward-time population genetic simulations of biophysically explicit gene models that can and do evolve sex-specific dominance reversal under sexually antagonistic selection, resolve sexual conflict, and maintain genetic variation. Among the insights gained from this effort thus far is that sex-reversed patterns of allele-specific expression are consistent with sex-specific dominance reversals for fitness. An allele-specific expression study lead by Prashasth Mishra has identified many such dominance-reversed candidate antagonistic genes in the fruit fly genome.
Sex-specific genetic architecture
Genetic architecture can can refer to either the mode of inheritance in, or the genetic covariances among, fitness and life history traits. These features ultimately determine a range of characteristics with basic and applied evolutionary implications such as the maintenance of genetic variation, the maintenance of sexual reproduction, adaptation, wildlife management, and public health.
I am broadly interested applying quantitative genetic methods to questions of the genetic architecture for fitness. Past quantitative genetics projects have used a variety of plant and animal systems to look at sex-specific dominance reversal, sex-specific selection against mutation load, the genetic basis of the male fighter/scrambler morphs, the (multivariate) genetic variance-covariance matrix (G), and the role of dominance in facilitating local adaptation. Looking forward, I am especially interested in connecting such methods with ‘omics methods in order to identify and characterize the underlying causal loci.